Hello! In this post, I want to give an elementary introduction into a topic that I have spent the last four years working on. I will be talking about a small topic in phylogenetics: species trees and gene trees. I will discuss what these trees are and why are they worth studying. I hope for this post to serve as an introduction to some more advanced topics I plan to cover later.
Let’s start by settling on terms and definitions. We will be talking about biological species and evolutionary relationships between them. In fact, whenever I say “species”, you can just think about a population of kittens/puppies/bunnies that is in reproductive and geographical isolation from other similar populations. New species are formed through speciation, a process in which one existing species splits in two. This can happen for many reasons, such as the appearance of a separating geographical barrier or ecological specialization. I will always assume that all of our species originate from a single ancestral species through a sequence of speciations.
Species trees
The history of the speciation events for a group of species is encoded by a tree, which we will call a species tree and denote by a letter \(S\). A species tree has nodes and edges. Each edge corresponds to a species that existed at some point in time. There are two types of nodes in \(S\): the tips of the tree are called leaves or leaf nodes, and the rest are called internal nodes. Each edge represents a species, with leaves (nodes and edges) representing the species that exist at present time. Each internal node represents some speciation event in the past.
Here is an example tree showing five species (called \(a\), \(b\), \(c\), \(d\), and \(e\)) descending from a common ancestor \(r\).

It’s time to meet a second protagonist of the story: a gene tree. To obtain a gene tree, we sample genetic data from a single individual in each species, and hand over the samples to the “experts”. They extract the DNA, sequence it, and then run bioinformatic algorithms with cryptic titles like CLUSTALW or MUSCLE to build a gene tree. Usually such gene tree is also given additional info such as branch lengths, but we don’t care about this for now. For us, both gene and species trees are purely combinatorial objects. Also, it’s important to note that we do not use any info about the species or the natural history behind them at any point in the process of building a gene tree.
Each edge of a gene tree corresponds to a genetic lineage, which is a chunk of DNA that is transmitted from parents to children through the generations. Each internal node of a gene tree corresponds to a coalescent event, in which an existing lineage splits into two due to some mutation appearing in one of the children lineages. A gene tree shows only those lineages and coalescence that we know about in our (tiny) sample.
Depending on your point of view, you can choose one of the two interpretations:
If you are into probability theory, you can think of the species tree as something that is known and fixed in the background, and the gene tree as the (random) data generated by a random process in which that species tree is a parameter.
If you are into statistics, you can think of the gene tree as a (known) observation, in which case the species tree is an unknown parameter that needs to be estimated.
But what is this random process I am talking about? And, more importantly…
Wait, shouldn’t the species and gene tree be the same?
It turns out that no, gene trees and species trees need not always be the same (have the same topology). Species trees operate at a macro level, dealing with populations, speciations, etc. — all are processes involving possibly thousands of individuals. A gene tree attempts to trace the genetic lineage throughout the past, and the result is dependent on randomness inherent in reproductive behavior.
The model that we have proposed to account for this is called the multispecies coalescent. In this post, I won’t try to explain what exactly is this thing. Instead, I will try to show you a single cryptic figure and let you figure it out yourself.
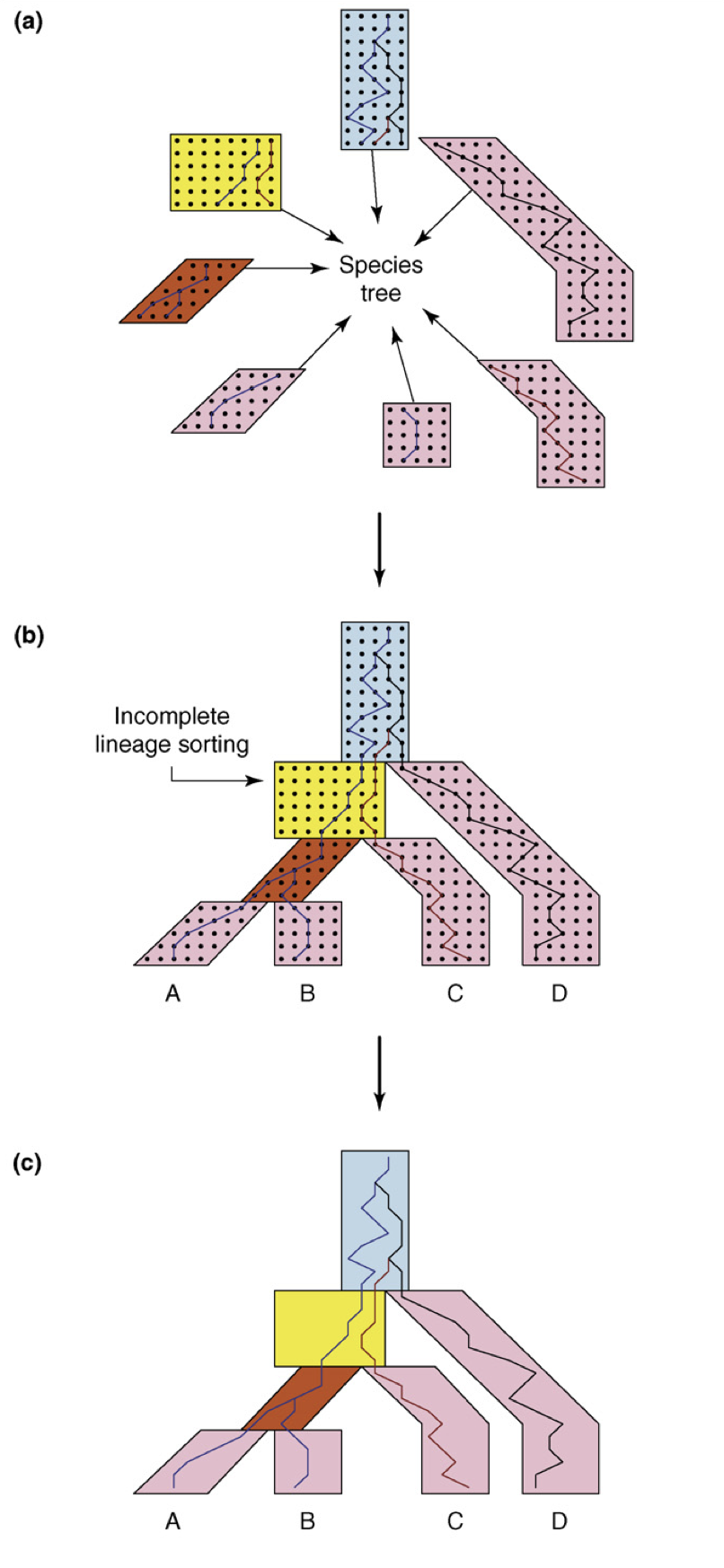
What I really want you to take away from this discussion is that, given any species tree, the multispecies coalescent can produce any gene tree topology with nonzero probability. This is the essence of it: macroscopic events such as the formation of the new species are reflected in microscopic changes in the DNA, and DNA is often the only thing we can observe! In thse cases, all of the inferences about the species’ natural history have to take into account this possible discordance of tree shapes! This means that we need to take seriously the study of the details of the the relationship between species and gene trees. That’s it for today!